Custom QC Algorithms
Mazama Science
2020-06-19
Source:vignettes/articles/Custom_QC_Algorithms.Rmd
Custom_QC_Algorithms.RmdData quality-control is the process of flagging or removing invalid or noise data points from a data set, either manually or algorithmicly. There are many statistical techniques and methods to preform quality-control. In this article we provide a general overview of how to construct such algorithms for use within the AirSensor package. Our goal is to write custom quality-control functions that replace, flag, or remove invalid data points. Data that are out of an instrument’s specifications, that appear to be associated with random noise or are associated with other known issues can be considered invalid.
NOTE: If you haven’t yet read the Temporal Aggregation
article, you should read it first in order to understand advanced usage
of pat_aggregate().
Example 1 – Count based QC
Let’s start by creating a simple QC algorithm based on the count of
the number of data records within an hour. If the count of records is
too low, we will replace any aggregated value with NA,
otherwise we will return the hourly aggregated value, in this example
the mean.
This QC function will create two hourly-aggregated ‘pat’ objects –
mean_pat and count_pat and use data from
count_pat to invalidate some records in
mean_pat.
## Loading required package: MazamaCoreUtils##
## Attaching package: 'dplyr'## The following objects are masked from 'package:stats':
##
## filter, lag## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
# Load a PurpleAir Timeseries ('pat') object
pat <- example_pat
# Define a QC function that returns an aggregated 'pat' object
QC_hourly_count <- function(pat, minCount = 11) {
# Create mean hourly aggregate pat
mean_pat <- pat_aggregate(pat, function(x) mean(x, na.rm = TRUE))
# Create count of non-missing pat data
count_pat <- pat_aggregate(pat, function(x) length(na.omit(x)))
# Create a minimum count mask
pm25_A_mask <- count_pat$data$pm25_A < minCount
pm25_B_mask <- count_pat$data$pm25_B < minCount
##################
# dplyr/tidyverse version
##################
#pat <- mean_pat
# mean_pat$data <-
# mean_pat$data %>%
# mutate(pm25_A = replace(.data$pm25_A, pm25_A_mask, NA)) %>%
# mutate(pm25_B = replace(.data$pm25_B, pm25_B_mask, NA)) %>%
# mutate(hr_count_pm25_A = count_pat$data$pm25_A) %>%
# mutate(hr_count_pm25_B = count_pat$data$pm25_B)
# pat <- mean_pat
##################
# base R version
##################
pat <- mean_pat
pat$data$pm25_A[pm25_A_mask] <- NA
pat$data$pm25_B[pm25_B_mask] <- NA
pat$data$hr_count_pm25_A <- count_pat$data$pm25_A
pat$data$hr_count_pm25_B <- count_pat$data$pm25_B
return(pat)
}
# Apply the QC function
hourly_count_pat <- QC_hourly_count(pat, minCount = 42)
# Examine the results
timeseriesTbl_multiPlot(hourly_count_pat$data, pattern = "pm25_[AB]")## Warning: Removed 336 rows containing missing values (`geom_line()`).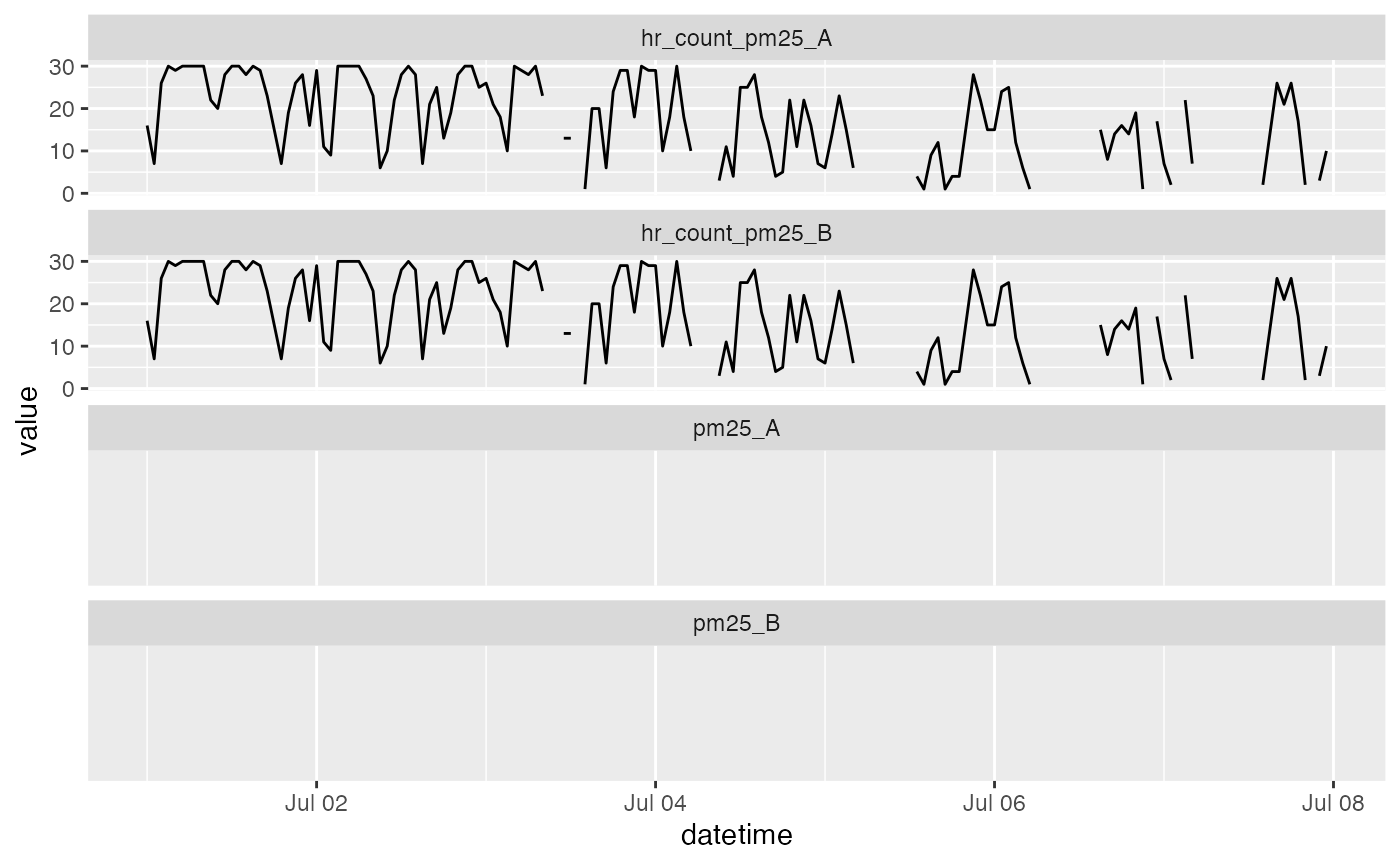
In the code above, we calculate hourly aggregated average values,
mean_pat, and hourly aggregated counts,
count_pat. We use the counts to create a mask identifying
hours where there is not enough data to meet our minimum threshold,
minCount = 42. The mean values for these hours are replaced
with NA.
Looking at the plot, we can verify that low count values are
associated with missing values of pm25_A/B.
Example 2 – Z-score based QC
In this example, we construct a more complex QC algorithm using
advanced features of pat_aggregate() and a coding style
appropriate for seasoned R developers.
The FUN parameter is defined “anonymously” (aka
on-the-fly) and calculates a Z-score for each 20-minute period in the
data. If the Z-score is below our 0.05 threshold, we
replace it with NA. The 20-minute data is then
re-aggregated to an 1-hour average and returned. Effectively, this
quality-control function is a basic two-tailed Z-score filter that only
aggregates data if the it is within the p-value (0.05).
QC_hourly_z_score <- function(pat, threshold = 0.05) {
pat_z_scored_20min_avg <-
pat_aggregate(
pat,
function(x) {
# Caluclate x z-scores
z_score <- scale(na.omit(x)) # (x - mean(x, na.rm = TRUE)) / sd(x, na.rm = TRUE)
# Create z-score mask -
# flag the z-scores who are out of the 5% and 95% quantile range
z_mask <- pnorm(-abs(z_score)) < threshold
x[z_mask] <- NA
# Calculate the 20-min average values without the flagged data
y <- mean(x, na.rm = TRUE)
return(y)
},
unit = 'minutes',
count = 20
)
# Re-aggregate to an 1-hour average of the data
pat <- pat_aggregate(pat_z_scored_20min_avg)
return(pat)
}
hourly_z_score_pat <- QC_hourly_z_score(pat)
# Examine the results
timeseriesTbl_multiPlot(hourly_z_score_pat$data, pattern = "pm25_[AB]")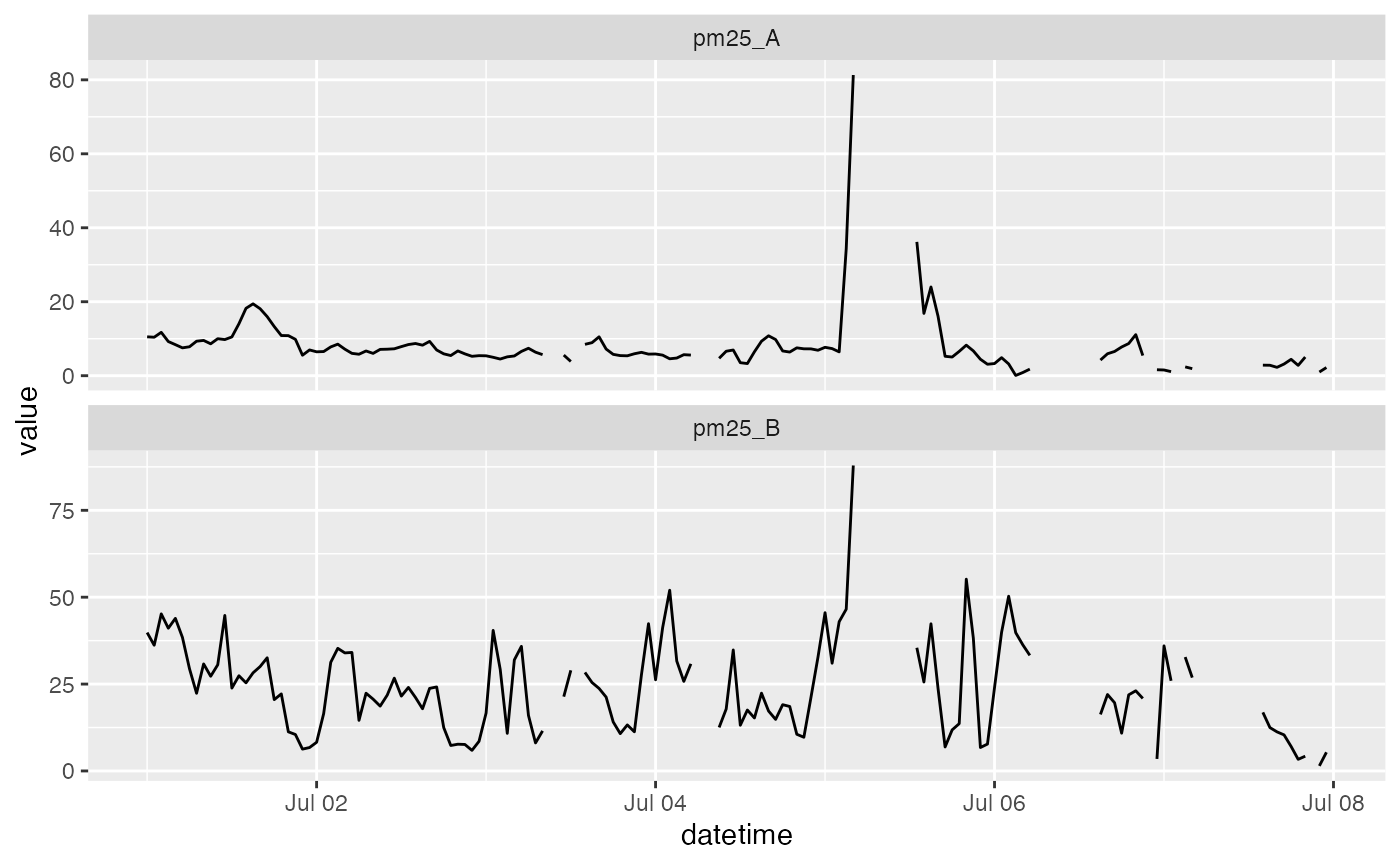
There are many ways to construct QC algorithms and the examples above illustrate what is possible within the AirSensor framework. We fully expect people to explore possibilities and create different QC algorithms depending on their individual needs:
- How important is it to remove all potentially bad data?
- How important is it to retain all potentially good data?
- Are there specific QC steps that required by an organization?
- …